Processing 1D Spectra in DNPLab
DNPLab is a Python package to process NMR data. The software was developed by Bridge12 and is open-source, free to use. DNPLab can be used for offline processing of OVJ data.
Below, a brief example is shown how to load and process a 1D NMR spectrum. For a complete reference and many more processing examples visit the DNPLab Online Documentation.
Installling DNPLab
DNPLab requires a working Python installation and can be simply installed using pip:
|
|
All requirements are automatically installed.
Processing 1D Spectra
A typical processing and plotting script is shown below:
|
|
The general structure of the script is:
- Line 1: Import the DNPLab package.
- Line 3: Load the NMR spectrum. DNPLab can import many different spectrometer formats. The format is automatically detected and a dnpdata object is created.
- Line 4: The attribute of the spectrum needs to be changed to nmr_spectrum to ensure that the plot function
fancy_plotworks properly (this line will not be required in the future). - Line 6-7: Fourier transform the spectrum and apply an autophase routine.
- Line 8: Remove DC offset from spectrum.
- Line 10-11: Plot the NMR spectrum
The result is shown below.
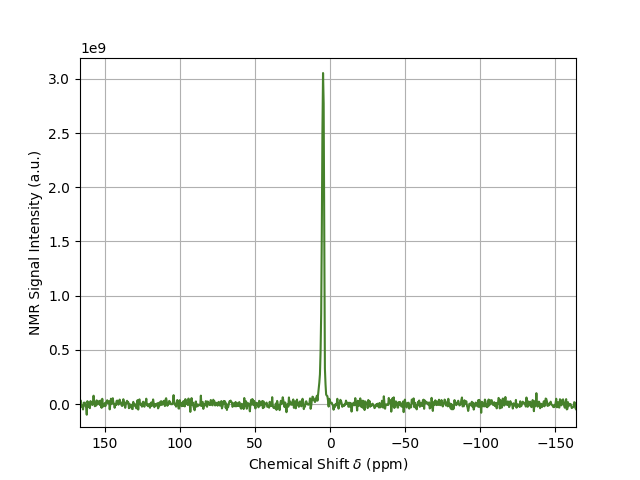
DNPLab has many more features for processing and analyzing NMR data. It can also be used to process EPR data. For more information check out the Online Documentation.