This section provides a brief overview how to acquire a 1D
- OVJ-B12 is installed on the computer
- The hardware is set up properly
- A sample is inserted into the probe
- The NMR probe is tuned to the spectrometer frequency.
Acquiring a 1H NMR
-
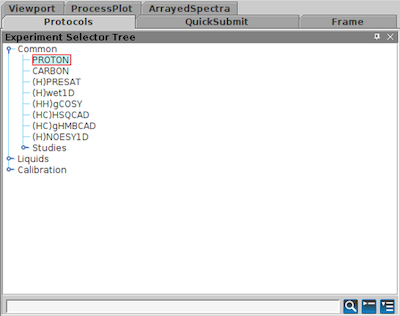
From the protocols tab drag the PROTON experiment into the plotting window, or double click on the PROTON protocol.
Alternatively, the pulse sequence can be set using the OVJ command line:
1seqfil='s2pul'The PROTON sequence calls a pulse program s2pul. This is a general, two-pulse sequence used to run variety of different experiments such as a single pulse NMR experiment or an inversion-recover sequence.
-
Next, the user has to set the length and power of the RF pulse to properly acquire a 1D 1H spectrum. In OVJ the pulse parameters can be changed from the OVJ command line or from the acquisition panel.
Note
Make sure that the spectrometer frequency is set correctly. The spectrometer frequency is set by adjusting the lockfreq. For experiments at 14.79 MHz, this parameter has a value 2.271 MHz (2H frequency corresponding to a 1H frequency of 14.78 MHz). This value should be ideally set to a precision of three to four significant digits.
See the Launching OVJ section in the Getting Started section for more details.
The length of the observe pulse is given by the parameter pw. From the OVJ command line set the pulse length:
|
|
corresponding to a pulse length of 3 µs.
Note
If the pulse length corresponding to a 90 degree pulse is unknown, it can be determined by performing a nutation experiment to calibrate the correct pulse length. Typically, the probehead manufacturer will provide the length and power level for the 90 degree pulse.
-
Next some of the acquisition parameters need to be set. Go to the Acquire tab and click on Acquisition. Here, basic acquisition parameters can be adjusted. Alternatively, the values can be entered in the OVJ command line.
- Spectral Width: Parameter sw
- Acquisition Time: Parameter at
- Complex Points: Parameter np
For example
1 2sw=50000 np=4096will set the spectral width to 50000 Hz (50 kHz) and the number of points acquired in the time domain to 4096.
Note
In OVJ the number of points corresponds to the total number of real and imaginary points. To acquire a total of 2048 (complex) points, np has to be set to 4096 (twice the number of complex points).
-
Set the number of transients (
nt) to 4. In general, OVJ will add a CYCLOPS phase cycle to each sequence automatically to remove DC offsets and correct for phase imbalances. Therefore, the parameterntshould always be set as a multiple of 4.1nt=4 -
On the Acquire tab click Channels to set the transmitter offset
tofparameter. To get started, set this value to 01tof=0With the transmitter offset set to 0, the spectrum, if excited on resonance, appears in the center of the window. To center the spectrum adjust the
tofvalue. Alternatively, you can click with the mouse on the peak (single red line cursor) and enter the commandmovetofin the OVJ command line. OVJ will automatically adjusttofso that the NMR spectrum in the next acquisition is at the center of the window. -
Start the acquisition by either pressing the GO button or by typing
1goin the OVJ command line. The acquisition will start and the data is displayed and processed on the screen.
Note
In OVJ the acquisition of the NMR spectrum can be started using the go or ga command. In this version of OVJ the two commands are identical. This may be different on other (older) NMR spectrometers running vnmrj.
Phasing a spectrum
Once the acquisition is finished, the spectrum most likely needs to be phased to show just the real part of the complex spectrum (see figure below). This can be done automatically using the autophase command aph or manually.
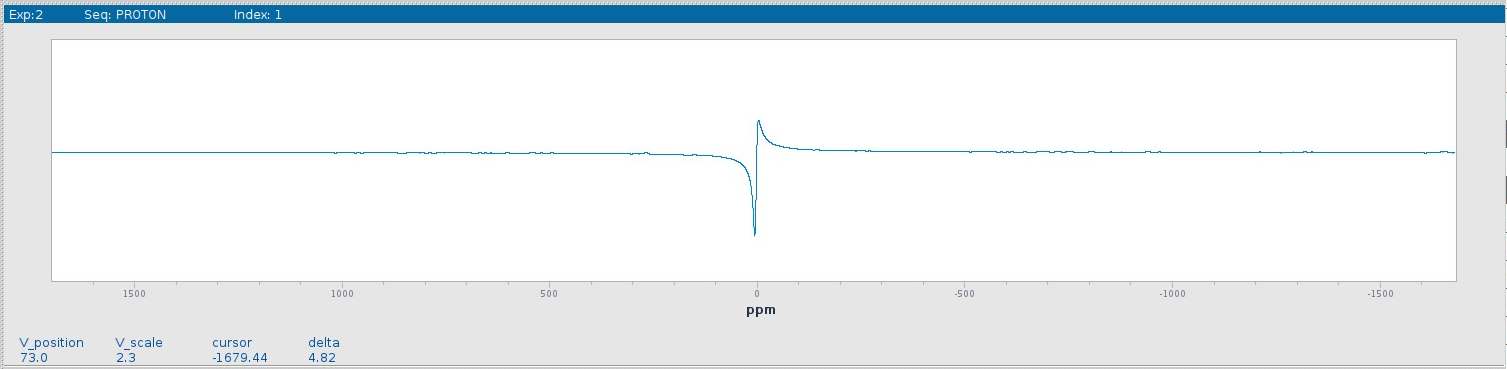
Using the Autophase Routine
To autophase the spectrum type the following command in the OVJ command line and press enter:
|
|
to autophase the spectrum.
Manually Phasing the Spectrum
Sometimes, the autophase routine fails and the spectrum needs to be phased manually. To manually phase an NMR spectrum:
-
Click the Phase button in the toolbar to the left

-
Click the spectrum with the left mouse button and keep the button pressed. Two red cursors appear to the left and right of the peak. Phase the peak by moving the mouse up and down. This will adjust the 0th order phase correction (frequency-independent phase correction). Alternatively, the phase can be adjusted from the OVJ command line by typing:
1rp=11.5 -
To adjust the 1st order phase correction (frequency-dependent phase correction) click the Phase button in the toolbar to the left. Next, select the peak by moving the mouse cursor to the NMR peak and click the right mouse button. The point selected is the pivot point used to adjust the 1st order phase correction. By moving the mouse up and down the phase of the rest of the spectrum can be adjusted. To adjust the 1st order phase correction
lpfrom the OVJ command line type:1lp=157
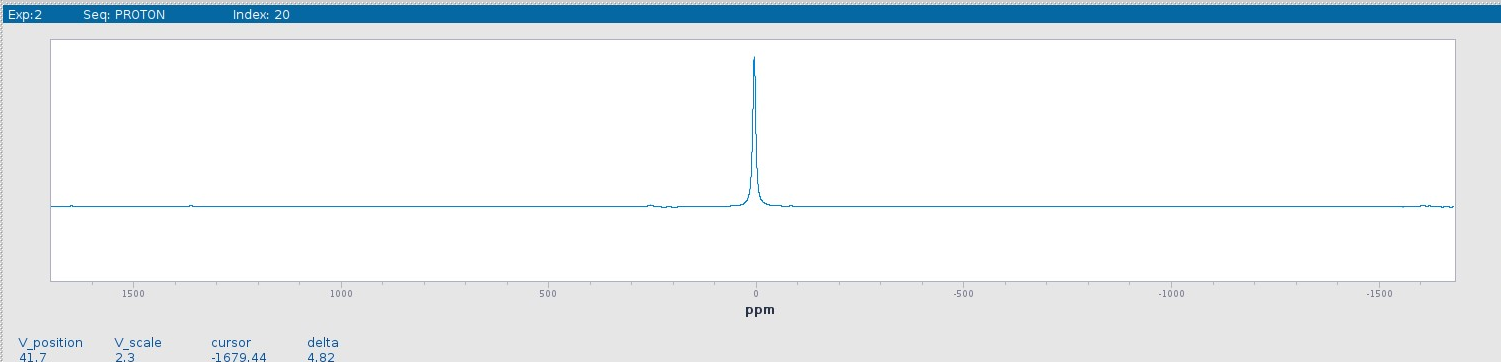
The proper phased spectrum is shown below.
Note
Start phasing the spectrum with rp and lp set to 0. First, adjust the 0th order phase and then the 1st order phase correction. Note that rp=360 is equivalent to rp=0. This is also true for lp.
Next, proceed to the next section to Save the NMR Spectrum.